Clear description for normalization method and DESeq2 normalization process (recommend)
https://hbctraining.github.io/DGE_workshop/lessons/02_DGE_count_normalization.html
Gene expression units explained: RPM, RPKM, FPKM, TPM, DESeq, TMM, SCnorm, GeTMM, and ComBat-Seq : include python, R code (recommend)
https://www.reneshbedre.com/blog/expression_units.html
calculation of CPM, TPM, FPKM ; R code (recommend)
https://haroldpimentel.wordpress.com/2014/05/08/what-the-fpkm-a-review-rna-seq-expression-units/
How to normalized TPM with TMM method?
https://www.biostars.org/p/388584/
->you have TMM factors and want to compute TPMs.
TMM factors can in principle be used to compute TPMs. In edgeR, any downstream quantity that is computed from the library sizes will incorporate the TMM factors automatically, because the factors are considered part of the effective library sizes. TMM normalization factors will be applied automatically when you use
CPM <- cpm(dge)
or
RPKM <- rpkm(dge)
in edgeR to compute CPMs or RPKMs from a DGEList object. I don't necessarily recommend TPM values myself, but if you go on to compute TPMs by
TPM <- t( t(RPKM) / colSums(RPKM) ) * 1e6
then the TMM factors will naturally have been incorporated into the computation.
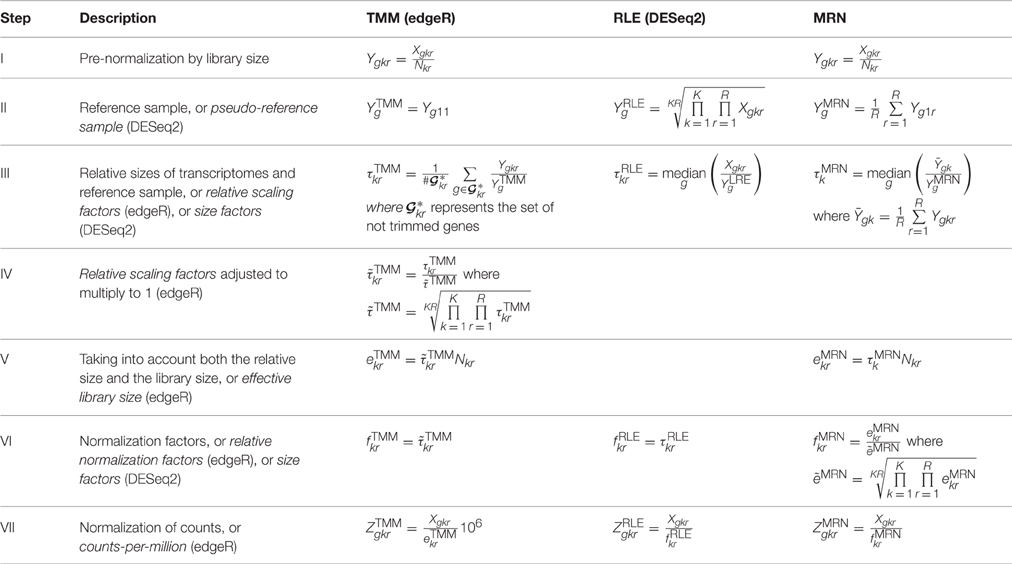
TMM (edgeR), RLE (DESeq2), and MRN Normalization Methods comparison
https://www.frontiersin.org/articles/10.3389/fgene.2016.00164/full
No comments:
Post a Comment