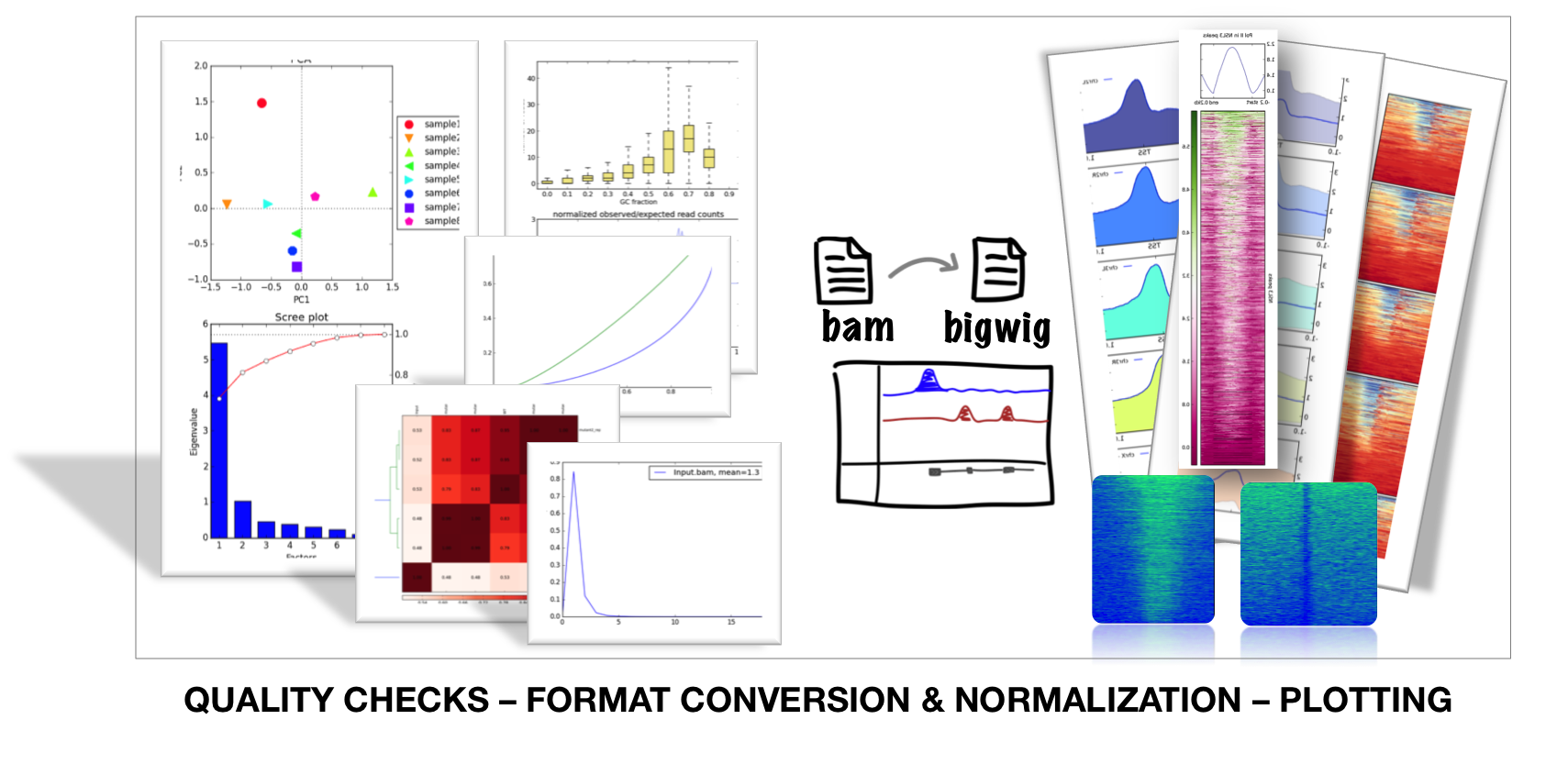
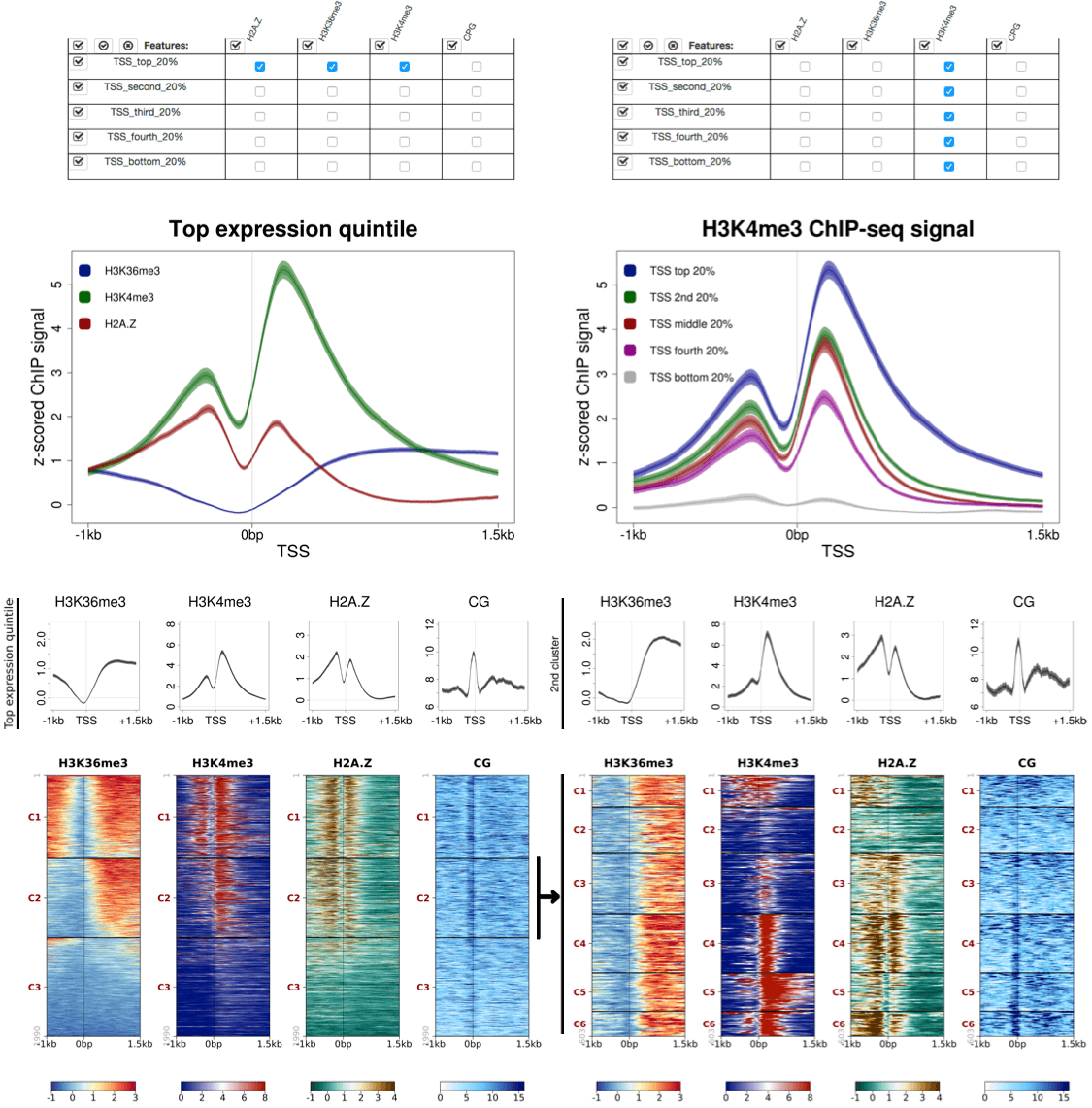
http://seqanswers.com/forums/showthread.php?t=22504
https://josephcckuo.wordpress.com/2016/11/17/modify-chromosome-notation-in-bam-file/
change chromosome notation "1" to "chr1"
for file in *.bam; do filename=`echo $file | cut -d "." -f 1`; samtools view -H $file | sed -e 's/SN:\([0-9XY]\)/SN:chr\1/' -e 's/SN:MT/SN:chrM/' | samtools reheader - $file > ${filename}_chr.bam; done
for loop
for file in *.bam; do filename=`echo $file | cut -d "." -f 1`; samtools view -H $file | sed -e 's/SN:\([0-9XY]\)/SN:chr\1/' -e 's/SN:MT/SN:chrM/' | samtools reheader - $file > ${filename}_chr.bam; done